Gao Lab
The Exposomics and Environmental Etiology (E³) Lab at Harvard T.H. Chan School of Public Health investigates the complex interactions between environmental exposures (collectively known as the exposome) and human health.
665 Huntington Avenue, Bldg 1, Room 311
Boston, MA 02115
Our Research
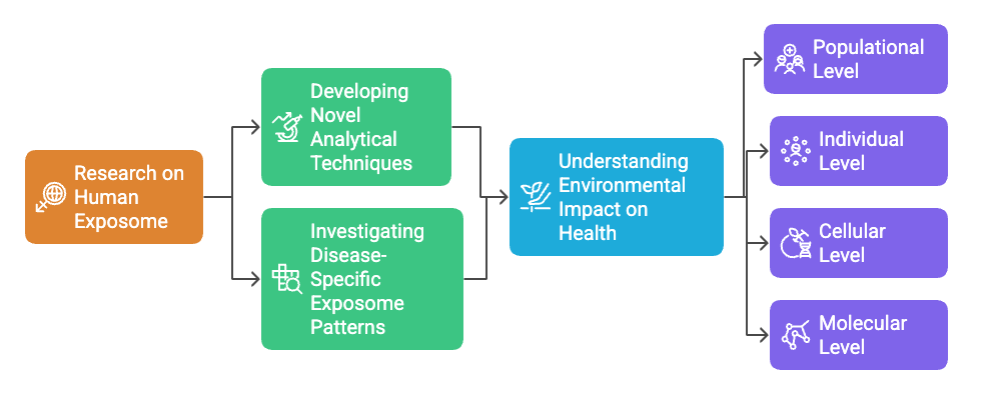
Our research spans multiple areas focused on decoding the human exposome and its impact on health outcomes.
From developing novel analytical techniques to investigating disease-specific exposure patterns, our work aims to provide a comprehensive understanding of how our environment shapes our health at the populational, individual, cellular, and molecular levels.
Exposome and Disease Etiology
We investigate the role of environmental chemicals in lung cancer development and progression, leveraging the Boston Lung Cancer Study and Pittsburgh Lung Screening Study cohorts. By examining blood chemical exposome and other omics profiles in lung cancer patients compared to controls, we aim to identify distinct environmental chemical signatures associated with increased risk. This work has potential implications for early detection, prevention strategies, and novel prevention approaches and therapeutic targets.
Our research explores how environmental exposures contribute to asthma severity and control. Using wearable environmental monitoring devices and biomonitoring approaches, we analyze the relationship between specific environmental contaminants and asthma-related biomarkers. This work informs our understanding of asthma pathobiology and could lead to improved prevention and management strategies.
We examine how specific environmental exposures influence the development and progression of Pulmonary Arterial Hypertension (PAH), particularly in idiopathic cases. Working with the University of Pittsburgh Medical Center PAH Cohort, we profile the blood exposome, metabolome, and proteome of PAH patients. This comprehensive analysis aims to identify environmental chemical signatures and elucidate their mechanistic roles through affected molecular pathways. Our work with this well-characterized cohort allows us to investigate PAH across various subtypes, including idiopathic, genetic, and systemic sclerosis-associated PAH. This integrative approach will lead to prevention approaches and therapeutic strategies for this complex cardiovascular condition.
We investigate the complex interactions between xenobiotics, the gut microbiome, and neurological health. Through our collaboration with the Florida Microbiome in Aging Gut and Brain Consortium Cohort, we analyze plasma samples with varying stages of cognitive function to identify exposome and metabolome signatures associated with cognitive impairment. Our research focuses on how gut microbial activity modulates xenobiotic and metabolite interactions that may promote neuroinflammatory and oxidative processes contributing to the progression of Alzheimer’s Disease and Related Dementias. This unique study allows us to explore the temporal relationships between exposures, microbiome changes, and cognitive decline, providing valuable insights into potential intervention targets.
Exposomics Methodology Development
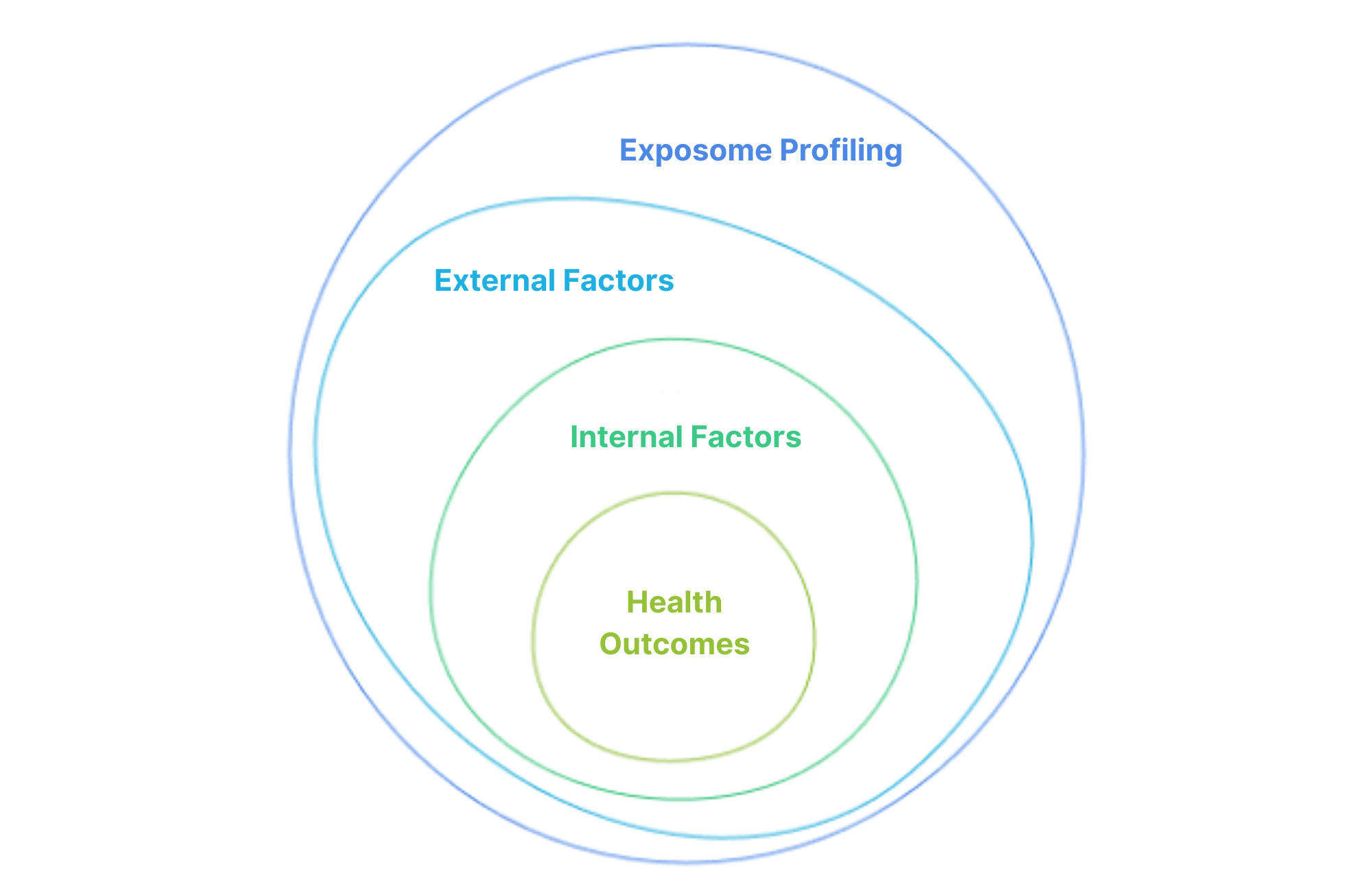
External and Internal Exposome Profiling
We develop and refine techniques for comprehensive monitoring of personal exposome profiles, creating innovative tools for continuous, non-invasive tracking of environmental exposures. Our approaches integrate organic pollutants, toxic metals, and airborne microbiota monitoring to provide holistic exposure profiles that reflect real-world conditions. Internal exposome profiling through biomonitoring measures chemical signatures in biological samples, connecting external exposures to internal body responses and establishing exposure-response relationships critical for understanding health impacts.
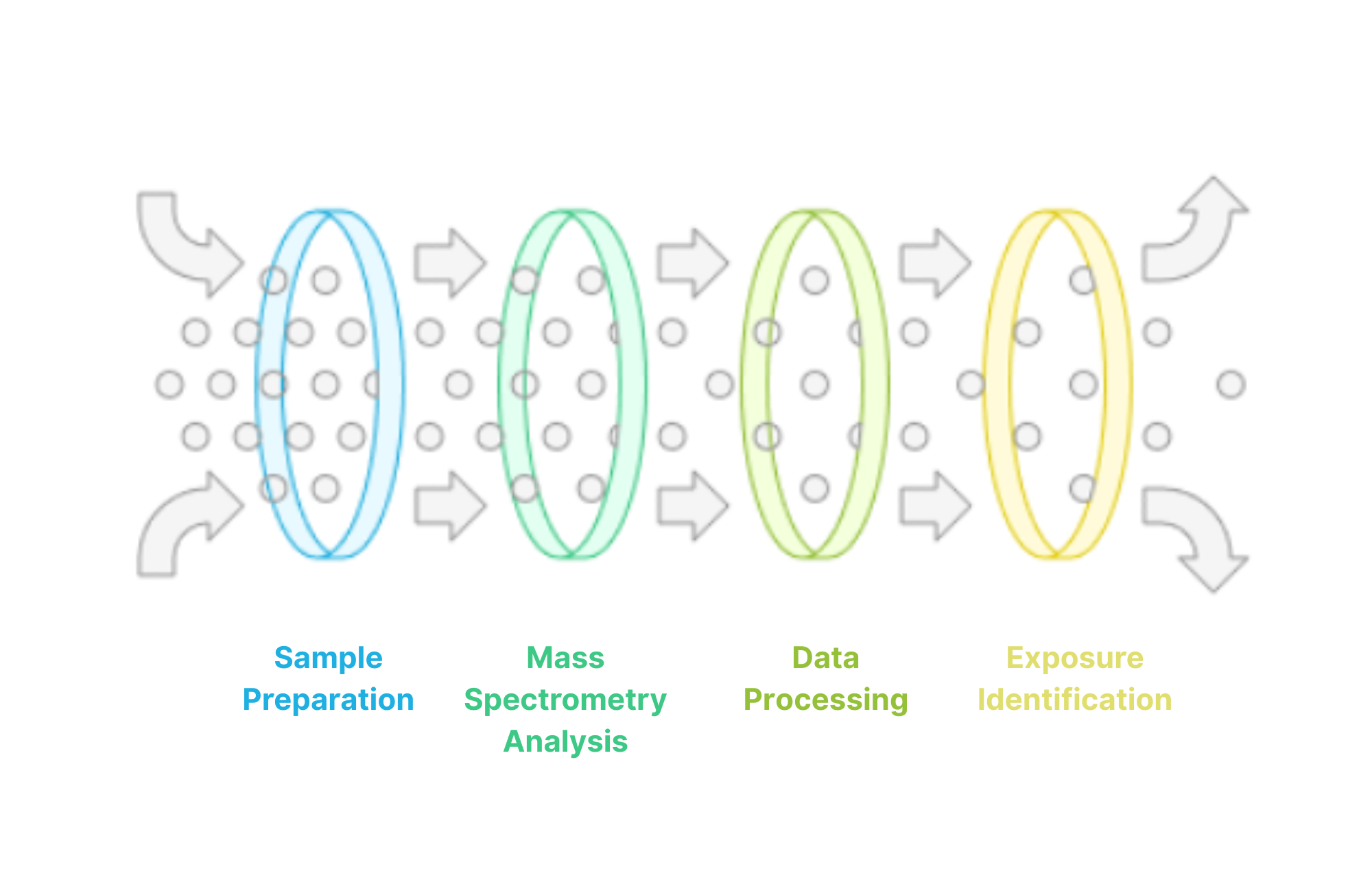
High-Resolution Mass Spectrometry-Based Exposomics
Our lab establishes high-throughput and integrative exposomics approaches using gas chromatography and liquid chromatography coupled with high-resolution mass spectrometry, as well as inductively coupled plasma-mass spectrometry. These techniques enable comprehensive identification/annotation and quantification of environmental chemicals in various matrices. We also pioneer innovative approaches to examine exposome profiles at the single/multi-cell levels, combining cell sorting with high-resolution mass spectrometry or imaging mass spectrometry. This cutting-edge research reveals how different cell types respond to environmental exposures, uncovering previously unknown vulnerabilities and adaptation mechanisms.
Emerging Contaminant Analysis
Our lab focuses on analyzing emerging contaminants of concern, including per- and polyfluoroalkyl substances (PFAS) and nano- and microplastics (NMPs). We develop innovative analytical techniques to detect, quantify, and characterize these emerging, persistent pollutants in environmental samples and biological matrices. By investigating their environmental fate, bioaccumulation patterns, and health effects, we aim to provide critical insights that inform regulatory decisions and public health policies. Our work on emerging contaminants bridges the gap between environmental monitoring and human health impact assessment, offering a comprehensive understanding of these challenging environmental threats.
Multi-Omics Integration
We integrate exposomics data with multiple omics layers (genomics, epigenomics, transcriptomics, proteomics, and metabolomics) by developing advanced bioinformatics pipelines to provide a comprehensive understanding of how environmental exposures influence biological processes and health outcomes. This integrative approach reveals complex relationships between external exposures and internal molecular changes comprehensively.
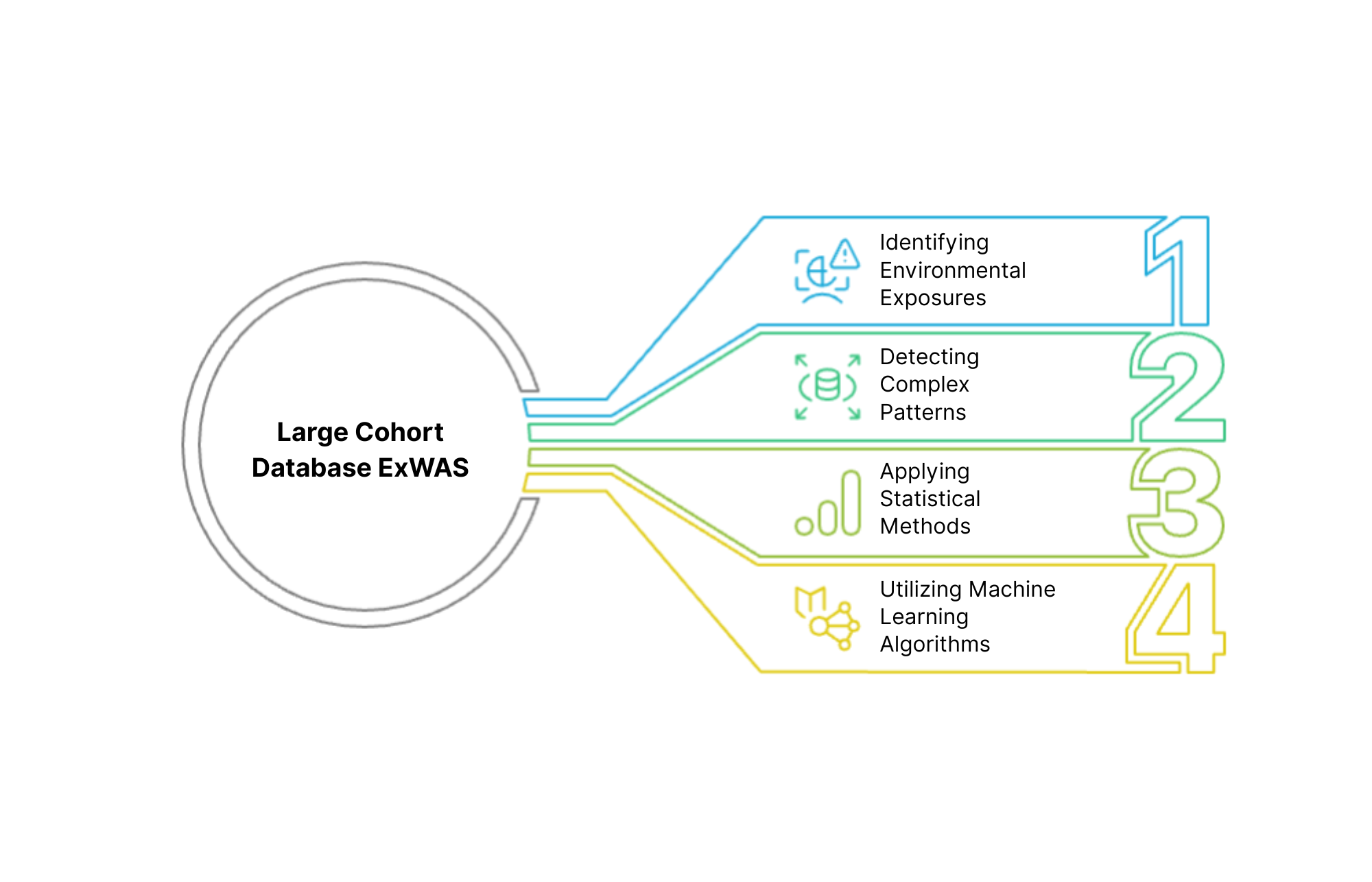
Large Cohort Database ExWAS (Exposome-Wide Association Studies)
We leverage large epidemiological databases and cohorts to conduct exposome-wide association studies, identifying relationships between environmental exposures and health outcomes at the populational level. By applying advanced statistical methods and machine learning algorithms to datasets such as the UK Biobank and All of Us, we can detect complex patterns and associations that might otherwise remain hidden.