I am Professor of Epidemiology with primary appointment in the Department of Epidemiology and a joint appointment in the Department of Immunology and Infectious Diseases, where my wet lab is located. I direct the Center for Communicable Disease Dynamics, a center of excellence funded by the MIDAS program of NIH/NIGMS. I am also the Associate Director of the Interdisciplinary Concentration in Infectious Disease Epidemiology.
Research (updating in progress)
My research concerns the effect of naturally acquired host immunity, vaccine-induced immunity and other public health interventions (e.g. antimicrobial use) on the population biology of pathogens and the consequences of changing pathogen populations for human health. Some of this work is motivated mainly by practical questions in public health (such as vaccine design and intervention targeting), and some is motivated by classical questions in population biology, such as how to explain patterns of coexistence of pathogen strains in space and time.
Studies of Streptococcus pneumoniae combine the practical and the population-biological questions, as well as the experimental and quantitative approaches.
Streptococcus pneumoniae: immune responses and population biology. Much of my work focuses on the bacterial pathogen Streptococcus pneumoniae (pneumococcus) which colonizes the nasopharynx of 30-100% of children worldwide, and causes otitis media, septicemia, pneumonia and meningitis in a small fraction, but a large number of them, with an estimated 800,000+ child deaths a year attributed to pneumococcal disease. Using experimental and epidemiologic approaches, we worked for over a decade on the question of how different serotypes of the pathogen coexist despite seemingly large fitness differences between different serotypes. That work began with a mathematical modeling study motivated by concerns about serotype replacement following the use of serotype-specific conjugate vaccines and with an experimental demonstration in mice that different strains of pneumococcus compete to colonize the upper respiratory tract. A series of studies in collaboration with the Malley lab at Boston Children’s Hospital showed that much of the acquired immune response to pneumococcal carriage was antibody-independent, and probably independent of complement but dependent on CD4+, Th17 cells and neutrophils. This immunity is independent of serotype and reduces duration of colonization rather than preventing acquisition. Work in this area in our laboratory was led by Krzysztof Trzcinski. We supplemented these experimental observations with epidemiologic analysis that showed evidence of modest serotype-specific immunity to colonization (collaboration with Ron Dagan‘s group), and acquisition of duration-reducing immunity to all serotypes in the first years of life (collaboration with Anthony Scott’s group including Osman Abdullahi).
Further work in the laboratory, led by PhD student/postdoc Daniel Weinberger, showed that multiple aspects of pneumococcal fitness were directly affected by serotype, such that certain serotypes with simpler capsular repeat unit structures and more negative surface charge have larger capsules, are more resistant to phagocytosis, are stronger competitors, and are more common in carriage. These relationships, shown in the figure, are summarized here and here.
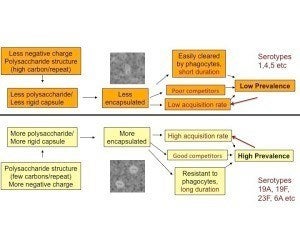
Given large fitness differences between serotypes and only weak serotype-specific immunity, how can there be so many serotypes; in particular how do the lower-fitness serotypes persist? In the course of studying this question, we realized that our (and others’) earlier mathematical models of serotype coexistence had inadvertently created “coexistence for free” in models by innocuous-seeming mathematical assumptions that corresponded to no known biological mechanism. Fixing that problem only made the problem harder.
Sarah Cobey took on the challenge of trying to explain serotype coexistence. She found that the combination of weak serotype-specific immunity (which enhances within-serotype competition) and duration-reducing, cross-serotype immunity (but neither alone) was capable of counteracting strong directional selection in favor of the “high-fitness” serotypes. Duration-reducing immunity, although not specific to individual serotypes, has a disproportionate impact on the long-duration (high fitness) serotypes because it reduces their duration of carriage proportionally more. We believe that these are the broad features of the answer to the question of how serotypes coexist despite strong directional selection.
We are now fitting this model to the existing data on serotype -specific carriage prevalence, with the goal of modeling the impact of existing conjugate vaccines and other proposed vaccines.
Pneumococcal population genomics and protein diversity. We have also been interested more broadly in the genomic diversity of pneumococci, and in particular in the diversity of protein antigens, some of which are targets of antibody elicited by the pneumococcus, and others of which are likely targets of CD4+ Th17 responses. Nick Croucher, Bill Hanage and I led analysis of 616 pneumococcal genomes from Massachusetts, collected 2001-7 by the SPARC project led by Jonathan Finkelstein and Grace Lee with Steve Pelton. Sequence analysis of isolates from later years is ongoing, involving the same team.
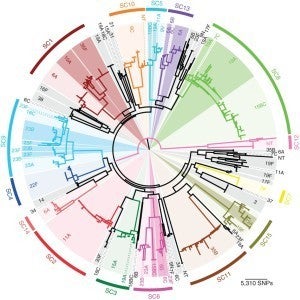
We are currently undertaking sequencing of a comparably-sized collection of pneumococci from Navajo and White Mountain Apache populations in the Southwestern US, in collaboration with Kate O’Brien of the Johns Hopkins Bloomberg School of Public Health. Bill Hanage is an active collaborator on this project.
Scientific questions of particular interest in population genomic studies include the role of immune selection in diversifying antigens other than the capsule in S. pneumoniae. We have shown age-specific patterns of gene presence for the pilus and some other proteins suggestive of immune selection. We have also shown, in work led by Yuan Li on a small genome sample that signals of diversifying selection are statistically apparent in proteins targeted by antibodies, and specifically in epitope regions. These signals are not present in proteins putatively targeted by CD4+ Th17 cells, perhaps reflecting the ability of such cells to prevent immune escape by acting in trans.
Current work in the laboratory focuses on elucidating the function of antiprotein antibody, the consequences of allelic diversity or target loss for bacterial fitness, and corroboration of these findings by comparing measurements of antibodies in human sera (collaboration with David Goldblatt) against pneumococcal genomes from isolates from the same humans at later time points. Modeling work (with Caroline Buckee) focuses on studies of the population-biologic effects of such immune responses.
Antimicrobial resistance in S. pneumoniae. Another problem in the population biology of pneumococci of longstanding interest is antimicrobial resistance. Early work focused on measuring the in vivo fitness cost of resistance and documenting the cotransfer of capsular and resistance loci in the lab. Quantitative studies considered the impact of antimicrobial use on resistance at the individual level and the interpretation of clinical trial data to assess this impact. Additionally, collaborating with colleagues at the CDC’s Active Bacterial Core surveillance, we have assessed trends in resistance at the national level and partitioned these into effects of changing serotype distribution and changing resistance within serotype, both before and since the introduction of conjugate vaccines.
A more basic question, still unsolved, is why drug-susceptible and -resistant strains of pneumococci coexist at the population level over long periods. An initial hypothesis that dynamics of resistance are intrinsically slow in the population has been refuted by analysis of the dynamics of resistance in Israel. A longstanding collaboration with Caroline Colijn, Christophe Fraser, Bill Hanage and Ted Cohen continues to explore alternative explanations for the phenomenon.
Applied pneumococcal epidemiology. In addition to these projects in population biology, we have a continuing interest in the applied epidemiology of pneumococcal carriage and disease. Projects have included collaboration on with SPARC, a visionary project led by Jonathan Finkelstein and Grace Lee with collaborators Bill Hanage and Steve Pelton , that provided a nearly unique view of the epidemiology of pneumococcal carriage through the introduction of two vaccines, PCV7 and PCV13. Studies have focused on the dynamics of serotypes and drug resistance, as well as clonal dynamics and invasiveness. Other work has included estimating serotype differences in disease severity, documenting serotype replacement in invasive pneumococcal disease globally (here and here) and investigating the extent to which carriage data can be used to predict invasive disease trends (here and here). Much of this work has been led by Daniel Weinberger, first while he was in our group and recently while he is on the faculty at Yale. We retain an interest in this area. Currently we are working with several groups to model the impact of pneumococcal vaccine programs.
Pandemic preparedness and response. With Megan Murray, James Robins and several other collaborators we made one of the earliest estimates of the reproductive number of the SARS virus during the spring of 2003, and later applied the same approaches to estimate the reproductive number of pandemic influenza in fall, 1918, showing it was around 2. This suggested the possibility of effective control measures, despite the difficulties inherent in controlling an illness in which transmission could precede symptoms. Working with Richard Hatchett and Carter Mecher, we showed that rapid, early nonpharmaceutical interventions in 1918 were associated with diminished spread of influenza. Other work has focused on the risk that multiple introductions could compromise attempts to contain a pandemic at the sources, and on the potential for drug resistance following massive antiviral use in a pandemic. Other work focused on how to define optimal targeting of scarce pandemic control measures, including antivirals and vaccines, by taking advantage of the data that may be available early in a pandemic (with Jacco Wallinga) or based on a transmission matrix (with Ed Goldstein and with Stephen Eubank’s group at Virginia Tech) or during the declining phase of an epidemic.
Response to the 2009 influenza A/H1N1 pandemic. With many collaborators, we were heavily involved in analyzing data and providing advice to public health authorities during the 2009 influenza pandemic. I served as a member of the 2009 H1N1 working group of the President’s Council of Advisors on Science and Technology (PCAST), and a co-author of its August 2009 report. I also served on “Team B” for the United States CDC, providing external advice during the pandemic. Early on, we published articles with international collaborators on decision-making under uncertainty during the pandemic and on how to maintain surveillance when cases become uncountable. We undertook several analyses of severity, first in Mexico, and later in the US and UK (with Anne Presanis and multiple collaborators in public health agencies). With Laura White and collaborators at CDC we made an early estimate of the pandemic reproductive number from CDC line list data. With Jeff Shaman, we correctly projected that no winter 2010 wave of the pandemic would likely occur in the US, apart from a small one in the Southeast. On questions of countermeasure optimization, we derived conditions for the predispensing of antivirals to high-risk persons to be life-saving and estimated the impact of targeting high-risk persons for vaccination. Karen Huang’s master’s thesis estimated a ~20% increase in the reproductive number of pandemic flu when schools opened in fall, 2009. We held an international meeting in 2010 to discuss lessons learned from the 2009 pandemic, resulting in an extensive summary report.
Antimicrobial resistance. We have worked on a variety of topics related to drug resistance in bacteria and viruses, with Bruce Levin, Carl Bergstrom, Megan Murray, Ted Cohen, and Matthew Samore as recurring collaborators. Because antimicrobial use affects not only the individual who uses it, but also the pathogen population, much of our effort has been to define the effects of antimicrobial use at the population level, focusing on S. pneumoniae drug resistance, resistance in hospital-acquired infections, among others. A recurring theme (in general, and in examples such as influenza and tuberculosis) is that optimizing treatment success for the individual host may simultaneously intensify selection for resistance in the population. We have shown (here and here) why efforts to cycle antibiotics to control resistance are unlikely to be effective. Other general phenomena of interest include the mechanisms of coexistence of drug-resistant and -susceptible organisms, the role of veterinary antimicrobial use in promoting resistance in human medicine, methods for analysis of antimicrobial resistance data, and the causes of multiple-drug resistance.
- MRSA: We have applied population genetic approaches to show that patient transfer between hospitals is associated with transfer of methicillin-resistant Staphylococcus aureus (master’s thesis of Weixiong Ke, collaboration with Susan Huang). Hsiao-Han Chang is currently updating this analysis with whole-genome data.
- N. gonorrhoeae: We have applied novel epidemiologic and population genomic analyses to data from the CDC’s Gonococcal Isolate Surveillance Program (GISP) to identify the role of sexual networks and multiple-drug resistance in the spread of ciprofloxacin-resistant gonococci (led by Ed Goldstein) and gonococci with reduced susceptibility to cefixime (led by Yonatan Grad). We are collaborating with Dr. Grad on further studies in this area.
Seasonal influenza: mechanisms and forecasting. The seasonality of infectious diseases is one of the oldest observations in medicine, yet the mechanisms underlying seasonality are poorly understood. Collaborations with Jeff Shaman to elucidate the role of absolute humidity in driving flu seasonality in temperate regions led to a correct prediction that pandemic influenza would not likely resurge in the US in winter 2010, except modestly in the Southeast. This has developed into systematic efforts at influenza forecasting centered in the Shaman Lab, which has taken leadership of this strand of research and which won the first CDC flu forecasting competition. We have also worked on predicting subtype abundance based on models incorporating inter-subtype competitive inhibition. We hope to design a randomized trial of humidification to reduce influenza transmission, and have collaborated on preliminary studies to that end.
Seasonal influenza: burden of disease estimation. We have developed and applied an improved regression framework to estimate the mortality burden from seasonal influenza, including multiple listed causes of disease. Talia Quandelacy‘s master’s thesis applied this approach to age- and sex-specific risks. Ed Goldstein, who led most of this work, is currently extending it to other outcomes including hospitalization. Jessica Jacobs’ dissertation estimated that ~13% of meningococcal disease in the US was flu-attributable.
HIV transmission modeling and behavioral aspects of STI transmission. A longstanding collaboration with the George Seage and the CEPAC group at Massachusetts General Hospital (Ken Freedberg, PI) has led to a series of projects using an individual-based model of HIV transmission, calibrated to South African HIV data using a Bayesian-melding-inspired approach, to study the potential transmission effects of HIV prevention interventions. The initial calibration effort, led by Alethea McCormick and Nadia Abuelezam is here, and further analyses are in progress.
Other work under development focuses on the detailed modeling of the “market” of sexual partnerships, specifically on how the number and makeup of sexual partnerships changes following behavioral interventions.
Mechanisms of vaccine action. When we use mathematical models to scale up vaccine effects on the individual to predict effects at the population level, we make strong assumptions about the how the vaccine alters an individual’s risk of infection over multiple exposures. The effect of challenge dose is typically ignored, even in infections where the exposure dose may vary over orders of magnitude, such as cholera. With Gabriela Gomes we have been exploring the consequences of modeling vaccines as shifting the dose-response curve to the right. An exciting collaboration led by Andrew Wargo, including Gabriela and Gael Kurath, examines this question experimentally using two vaccines against diseases of salmon. We are also interested in the statistical problems of estimating vaccine effects in trials (two separate chapters of Justin O’Hagan’s doctoral thesis) and the problems of incorporating vaccines into models of transmission dynamics.
Development of new methods for infectious disease data analysis. We frequently develop new methods to deal with novel types of data related to infectious disease. Examples include:
- MCMC methods for assessing serotype replacement vs. unmasking in pneumococci and
- MCMC approaches to analyze household data to assess time-varying infectiousness with SARS (with Virginia Pitzer and Gabriel Leung)
- Hidden Markov models for hospital-acquired infections (with Ben Cooper) and new approaches for analyzing data on antimicrobial use and resistance (dissertation work of Sibel Ascioglu). The common thread here is accounting for the dependence of incidence on prevalence.
- An approach to quantify the tradeoffs of sensitivity, specificity and timeliness in epidemic alerts, applied to highland malaria in Ethiopia (with Hailay Teklehaimanot).
- Use of temporal variation in risk factors for infectious disease to infer transmission networks. This began with a collaboration with Jacco Wallinga on intervention targeting based on age-specific incidence patterns, and with Virginia Pitzer‘s innovative etiologic investigations of Kawasaki disease based on seasonal variability in mean age of incidence. More recently, Ed Goldstein and Colin Worby have been using changes in the age-specific incidence of several diseases to infer aspects of the transmission matrix.
- Definition of a class of “neutral null models” to improve the study of multi-strain pathogens (with Caroline Colijn, Christophe Fraser and colleagues)
- Causal definition of the per-contact efficacy of vaccines (Justin O’Hagan‘s dissertation)
- Novel uses of syndromic surveillance and symptom profiles to estimate incidence of specific infections (led by Ed Goldstein with Oscar Patterson-Lomba)
Population Genomics. A growing focus of our group has been on the population genomics of pathogenic bacteria and viruses, with a focus on how population genomic analysis involving epidemiologic “metadata” can enhance public health. Projects include:
- Whole-genome sequencing of E. coli O104:H4 isolates during the German/French outbreak of 2011 to assess possible epidemiologic scenarios and place it in the context of other outbreaks (led by Yonatan Grad, collaboration with Bill Hanage and the Broad Institute)
- Longitudinal eep sequencing of respiratory syncitial virus (led by Yonatan Grad) in a single immunocompromised patient to assess immune responses’ effect on viral diversity
- Ongoing analysis of the evolution of Streptococcus pneumoniae in Massachusetts (see section above on S. pneumoniae population genomics); analysis of the first 616 isolates from 2001-7 was published in 2013.
- Ongoing analysis of the genomes of ~800 isolates of pneumococci from Navajo and White Mountain Apache populations in the Southwestern US, in collaboration with Kate O’Brien of the Johns Hopkins Bloomberg School of Public Health. Bill Hanage is an active collaborator on this project.
- Ongoing analysis of 12 years of Bordetella pertussis isolates from Massachusetts to assess vaccine impacts on the population structure, led by Colin Worby with Bill Hanage.
- Ongoing analyses of the population genomics of antimicrobial-resistant Neisseria gonorrhoeae, led by Yonatan Grad (begun while he was a postdoc, continuing now that he is a faculty member in our Department of Immunology and Infectious Diseases). The first output from this project was a genomic analysis of gonococci with reduced susceptibility to cefixime in the US. Continuing work involves several more detailed samples of local epidemics, as well as further analysis of national samples.
- Methodological work on the inference of transmission from pathogen sequence data and the distribution of pathogen genetic distances in transmission networks (led by Colin Worby, co-supervised by Bill Hanage).
Science Policy: Creating Potential Pandemic Pathogens (PPP) in the Laboratory. I have been active in the debate over potential pandemic pathogen creation in the laboratory, arguing that the creation of novel, transmissible virulent influenza strains is unsafe, that the benefits are overstated, and that alternative, safe scientific approaches can provide comparable or greater public health benefits with minimal risk. This discussion has been active since the publication of two papers in 2012 by the Kawaoka and Fouchier laboratories. At the time, colleagues and I pointed out the limitations and risks of such experiments, with a subsequent collaboration with Alison Galvani focusing attention on ethical aspects and alternative approaches, and on the barriers to achieving the experiments’ claimed public health goals. Attention was refocused on laboratory safety by laboratory mishaps in spring-summer, 2014 at the NIH and CDC, which I argued in a New York Times op-ed should stimulate careful risk assessment before such work proceeds. In July 2014 16 scientists cofounded the Cambridge Working Group, which called for PPP experiments to be curtailed pending a risk-benefit assessment, a call that garnered over 300 signatures from prominent scientists and others. On October 2014 the White House announced a 1-year funding pause on such experiments, pending a risk-benefit assessment. Recent efforts have included an outline of the calculations that should enter such an assessment (with Tom Inglesby) and pieces for more general readers including a debate in Nature Reviews Microbiology and an opinion piece Scientific American. In progress are further explorations of the ethical dimensions with Nick Evans. My Reddit AMA on the topic is here. I recently co-founded the Society for Safe Science with Marcel Salathé. A 2015 debate on the topic at the Cambridge Center for the Study of Existential Risk with Derek Smith lays out the major issues.
Education
B.A., 1991, Philosophy, Yale University
D.Phil., 1995, Zoology, University of Oxford
Photo Credit: Stephanie Mitchell/Harvard University News Office



